Dynamo uses flow and pressure data to provide localised ‘areas of interest’ following a burst occurring. We have recently developed a new version of the system which implements the analysis in such a way that we can detect and localise burst events live throughout the day, as soon as the relevant flow and pressure data becomes available. This allows Dynamo to always be looking for new events and ensure our clients get insight as soon as possible.
When implementing a live system, the speed of processing becomes essential to its success. If we can’t import, analyse and report results from the data faster than the data is coming in, then it won’t work as a live system!
One of the ways in which we’ve optimised the performance of Dynamo, is by adopting concurrency where possible. Concurrency in this context means doing more than one thing at the same time, with the goal of improving performance by maximising the utilisation of server resources (e.g. CPU usage).
This blog will use examples of how multiprocessing has been used to speed up the time series data import to provide insight into how this can be achieved in Python. Rather than giving a comprehensive tutorial (of which several are available elsewhere), the aim here is to share some of our experiences in utilising the Python multiprocessing library and give an example of how it can be used to optimise the performance of some tasks.
Background
Firstly, a brief introduction to concurrency, in which some concepts have been simplified to make them more palatable and fit them in to this short blog. It was mentioned in the introduction that the method aims to maximise the utilisation of resources. There are two approaches to this:
- Multithreading
- Multiprocessing
In this post, we will be talking exclusively about multiprocessing.
A computer’s CPU has one or more processor cores, each of which can be thought of as being able to work on one task at a time, which it calls processes. Modern computers normally have multi-core processors on which we should be able to work on multiple tasks in parallel.
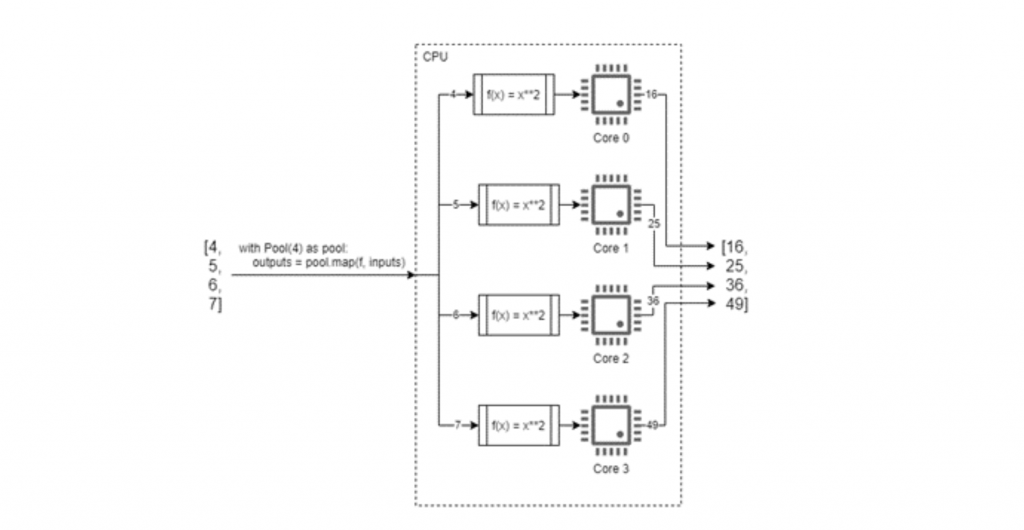
Your computer appears to be running hundreds of processes to run the applications you have open but in reality, each of its cores is only running one process at a time. It multitasks by switching between applications at opportune moments. However, using Python’s multiprocessing library, we can achieve true parallelism. We can apply this when performing the same operation many times over similar inputs, allowing us to improve performance.
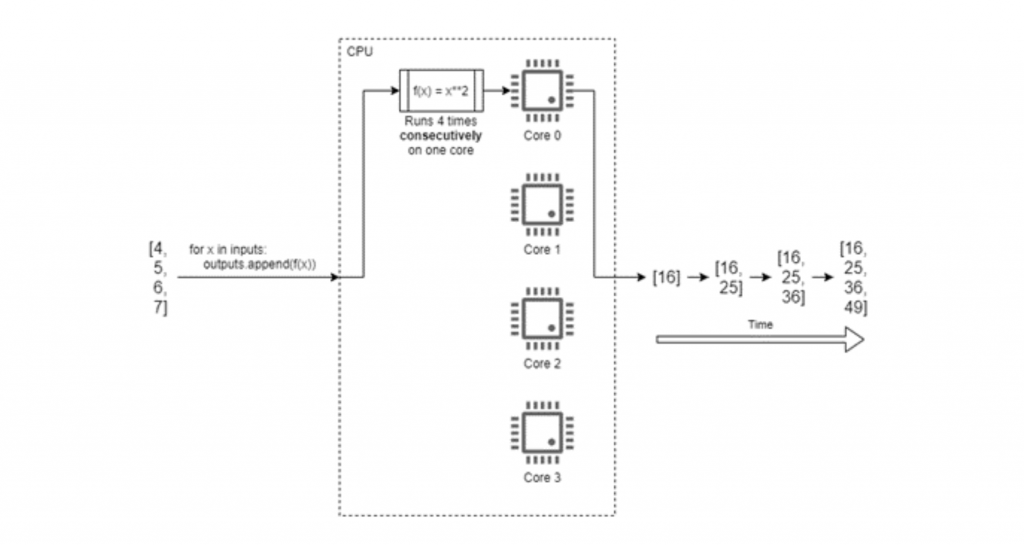
For example, consider that we have a list of 4 numbers which we want to square. A program to solve this will run so fast that we don’t need to optimise its execution time, but it serves as a good example. A conventional approach might be to:
- write/appropriate a function which accepts a single number as an argument and returns the square of that number; and
- write some procedural code which uses a for loop to iterate over the list, executing the function on each element and appending the results to a second list.
Figure 1 shows how Python would execute this code on a quad-core computer. The running time of our program will be about 4 times as long as it takes to calculate the square of one number, as the numbers are squared one-by-one. We cannot start calculating the square of the second element before we’ve calculated the first, and so on.